
 English
English
 French
French
The genetic factors in asthma
Les facteurs génétique dans l’asthme
H. Nguyen-Thi-Bich1, T. Van-Khanh2, H. Le-Thi-Minh1, S. Duong-Quy3,4,5
1: Department of Asthma and Immuno-Allergology. National Hospital of Pediatrics. Hanoi, Vietnam
2: Center of Gene and Protein Research. Hanoi Medical University. Hanoi, Vietnam
3: Cochin Hospital. Paris Descartes University
4: Penn State Medical University
5: Lam Dong Medical College. Dalat, Vietnam
Corresponding author
Pr. Sy DUONG-QUY.
Hôpital Cochin. Paris, France. Université Penn State.USA
Collègue de Médecine de Lam Dong. Dalat, Vietnam.
E-mail: sduongquy.jfvp@gmail.com
ABSTRACT
Asthma is a disease with diversified clinical and pathophysiologic mechanisms, characterized by the chronic inflammation of the airways, bronchial hyperresponsiveness, and reversible of the airway obstruction. Pathophysiologic features of the asthma is expressed in the interaction between genetic and environmental factors. Genetic factors in asthma were discovered about three centuries ago when the scientists found that 33-77% of genetic asthma rate was estimated in the study done on twins among one group of families. Although the majority of patients have good treatment response to 3 major groups of asthma drugs, including beta2 agonist, corticosteroids, anti-leukotriens, a small number of patients do not respond. Corticosteroid works effectively in treating asthma, its side effects such as asthma children’s growth limitation, bone density reduction, adrenal insufficiency, causing the anxiety to patients, their family and doctors in clinical practice.
Therefore, it is increasingly concerned to understand the genes that predict the corticosteroid response, which will help patients’ treatment accurate, effective, and avoid the unwanted side effects, especially in children. Genetic studies of asthma showed a very important role of genes in asthma because it also helps to to classify asthma phenotypes and to predict the medication response to personalize asthma treatment.
KEYWORDS: Asthma; genetic factors; corticosteroid response; asthma phenotype.
RÉSUMÉ
L'asthme est une maladie avec des mécanismes cliniques et physiopathologiques diversifiés, caractérisée par l'inflammation chronique des voies aériennes, l'hyperréactivité bronchique et la réversibilité de l'obstruction des voies aériennes. Les caractéristiques physiopathologiques de l'asthme sont exprimées dans l'interaction entre les facteurs génétiques et environnementaux. Les facteurs génétiques dans l'asthme ont été découverts il y a environ trois siècles lorsque les scientifiques ont constaté que 33-77% du taux d'asthme génétique a été estimé dans l'étude faite sur les jumeaux dans un groupe de familles.
Bien que la majorité des patients ont une bonne réponse au traitement à 3 grands groupes de médicaments contre l'asthme, y compris l'agoniste de bêta 2, les corticostéroïdes, anti-leucotriènes, un petit nombre de patients ne répondent pas. Corticostéroïde marchent efficacement dans le traitement de l'asthme, ses effets secondaires tels que la limitation de la croissance des enfants asthmatiques, la réduction de la densité osseuse, l’insuffisance surrénalienne, causant l'anxiété aux patients, à leur famille et aux médecins en pratique clinique. Par conséquent, il est de plus en plus préoccupé de comprendre les gènes qui prédisent la réponse des corticostéroïdes, ce qui aidera le traitement des patients précis, efficace et éviter les effets indésirables, en particulier chez les enfants. Les études génétiques ont montré un rôle très important dans l'asthme parce qu'il aide aussi à classer les phénotypes d'asthme et à prédire la réponse de médicament afinde personnaliser le traitement de l'asthme.
MOTS CLÉS: Asthme; facteurs génétiques; réponse aux corticostéroides; phénotype de l’asthme.
BACKGROUND
Asthma is a disease with diversified clinical and pathophysiologic mechanisms, characterized by the chronic inflammation of the airways, the increase of bronchial response and spasms [1]. Pathophysiologic features of the asthma is expressed in the interaction between genetic and environmental factors [2].
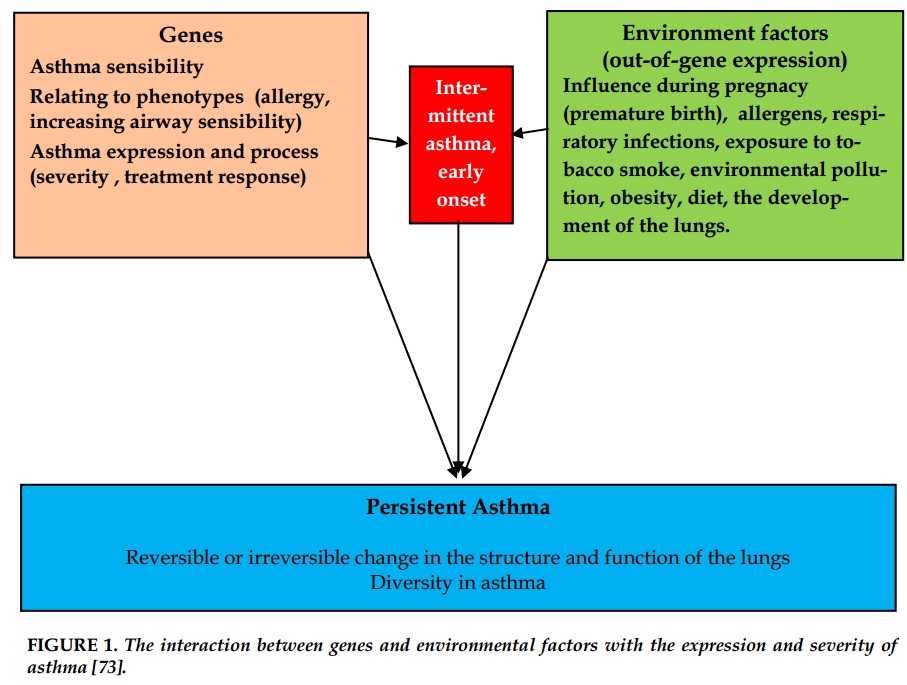
Asthma is a multi-gene and polymorphism (phenotype) disease, so the interaction with environmental factors plays a very important role in the study of genetic factors in Asthma. However, until now, the genetic phenotype specifically in asthma has not been clarified.
Thus, the aim of studying the genetic factors in asthma is not only to determine the susceptibility of the human body to the disease, but also to evaluate the progress and severity of the disease through genetic analysis. The results of GWAS (Genome-Wide Association Study) recently announced help clinicians better understand the genetic characteristics of asthma.
Genetic factors in asthma were discovered about three centuries ago when the scientists found that 33-77% of genetic asthma rate was estimated in the study done on twins among one group of families [3-7]. Until 1983, more than 190 genes involved in asthma heredity were published in over 1000 papers; and so far, the number of genes involved in asthma has been increasing [8-9]. The genetic factors in asthma is not due to a specific gene, but countless collections of genes; therefore, asthma gene phenotypes are various and treatment response varies on each patient.
Although the majority of patients have good treatment response to 3 major groups of asthma drugs, including beta2 agonist (or stimulating beta 2), corticosteroids, anti-leukotriens, a small number of patients do not respond. According to previous studies, 60-80% of genetic factors contributes to the treatment response of individuals [10].
Corticosteroids are first-line medication for the treatment and prevention of asthma in children and adults as recommended by GINA for its effective anti-inflammatory effects. But the response to corticosteroids is different in each patient and the genetic factor is one of the reasons leading to this different response.
Among the genes related to corticoid response, FCER2 and CRHR1 have been mentioned regularly in several studies. CRHR1 (corticotropin - releasing hormone receptor 1) is a gene coding the receptor involved in releasing the adrenocorticotropin hormone (ACTH) and conditioning the cortisol levels in blood. CRHR1 has a key role in the way to respond to corticoid. Mutations of replacing G by T in rs242941 of CRHR1 relates to the improvement of gene, which plays an important role in the process of bio-synthesis and activity of IgE (conditioning the reverse inhibition of the IgE synthesis and activity). Studies have shown that the mutations of replacing T by C in rs28364072 of FCER2 relate to asthma attacks and hospitalization times of patients inhaling corticosteroids [13-14].
Although corticosteroids works effectively in asthma treatment, its side effects such as asthma children’s growth limitation, bone density reduction, adrenal insufficiency, causing the anxiety to patients, their family and physicians in clinical practice. Therefore, it is increasingly concerned to understand the genes that predict the corticoid response, which will help patients’ treatment accurate, effective, and avoid the unwanted side effects, especially in children.
Characteristics and genetic analysis methods in asthma
Genetic characteristics of asthma
By far, allergic diseases like asthma are known for genetic characteristics of individuals in the same family. In 1916, a study of 621 people with atopy and 76 without atopy showed that 48.43% of the former group got allergy while this rate of the latter group was only 14.4 % [15]. Similarly, the study of 176 families showed that there was a close relationship between asthma parents and children suffering asthma and eczema [16]. Pairs of twins are often studied to determine genetic characteristics in asthma. A large study of 11,688 twins in Sweden showed that 73% of sensitivity in asthma is due to genetic factors and under the interaction with environmental factors [17].
Asthma has genetically complex disorders with genetic characteristics. This complex disorder inherites across generations clearly but does not follow the laws of Mendelian genetics in single gene diseases such as Huntington's syndrome, cystic fibrosis or diseases caused by recessive mutations inherited in line of soma.
Asthma heredity is the combination of changing various genes under the interaction with different environment factors forming varied phenotypes.
Characteristics of the multifactorial disorder is that patients depend on the level of heredity interactions of genes linked to disease. Therefore, unlike the single-gene genetic diseases, the expression level of asthma patients is very diverse, various and not in a linear relationship. This makes asthma prediction and prevention more difficult [18].
The important impacts of environmental factors on the expression of asthma include the exposure to tobacco smoke, respiratory infection, diet and exposure to respiratory allergens. Besides, gender and race also have an important role in the expression of asthma. There were many studies to evaluate the interaction of genetic factors with the exposure to tobacco smoke and risk of asthma. The results of the cohort study on the association between pregnancy or exposure to passive tobacco smoking of newborns with genetic abnormalities and risk of asthma have been demonstrated.
The role of genetics in clinical phenotype
GWAS studies have identified genes that predict asthma, relating to asthma onset period and the genes involved in allergic asthma phenotype, the severity of asthma, the increase of bronchial reaction.
Genes involved in allergic phenotype
Eosinophils is a biological marker related to the inflammatory response in asthma. A GWAS study in 9392 people of Ireland and some studies in Europe and East Asia showed that the gene polymorphism of IL-1RL1, WDR36, IL-33, MYB related to eosinophil numbers and allergic asthma [42].
Genes related to disease severity
According to the research by Li et al. in 473 Hispanic patients and 1.892 other patients in the study of TENOR (The Epidemiology and Natural History of Asthma: Outcomes and Treatment Regimens), genetic variations of RAD / IL13 in 5q31.1 position relate to the severity of allergic asthma [43-45]. ADAM 33 gene (A disintegin and metalloprotease 33), a member of genes relating to adhesion, transmiting the signal in cells, using position merchandise flow method, define that this gene get involved in asthma. In addition, in the correlation study, ADAM 33 relates to patients’ respiratory functions, thus, relates to the severity of asthma. ADAM 33 gene polymorphisms predict diminished indicators of FEV1, concerning the asthma process. In a study of 1.441 patients with asthma, geneti polymorphism of HHIP/rs1512288 on chromosome 4q31 relates to FEV1 and FVC index. HHIP gene is also linked to the bronchi recovery but not to the increase of the bronchial response [46].
In a study of SARP (Severe Asthma Research Program) in the US, genetic polymorphisms related to IL-4 receptor are IL-4 E375A and Q551R related to severe asthma attacks and poor respiratory function, and E375A C allele relates to the increase of mast cells [47]. Asthma phenotypes were divided into 5 clusters, according to SARP, including Cluster 1: mild allergic asthma; Cluster 2: mild-average allergic asthma; Cluster 3: late-onset allergic asthma; Cluster 4: severe allergic asthma; Cluster 5: severe asthma with airway obstruction [48]. Clusters of 1, 2 and 4 have large percentage of families with a history of allergy compared with Clusters of 3 and 5, while the related genes of pathway IL 4/13 towards Th2 are more prevalent in the Cluster of 1, 2 and 4 [49].
Genes related to the susceptibility status of airways
Koppelman studying 200 Danish families found that the gene polymorphism of PCDH1 at the position 5q31.3 related to airway hypersensitivity [24]. In the study by White et al, OPN3 genes at position 1q43 and CHML at 1q42-qter related to allergic asthma and airway hypersensitivity [50].
In summary, the study of sensitive genes in asthma has achieved positive results, but the pathogenesis of severe asthma as well as an understanding of the gene impact on the severity of the asthma is limited. Therefore, it is important to determine genes involved in asthma severity, in respiratory functions, in the increase of bronchial reactions in order to help diagnose asthma phenotype groups for finding effective and appropriate medication in each asthma phenotype.
The role of heredity in the asthma pathogenesis
The studies of basic genes in asthma brought impressive knowledge in the pathogenesis of this complex disease. Initially, most studies on gene candidates of asthma focused on the connection of functional polymorphisms in the components of the immune response by Th2. Cytokines-encoding genes through Th2 is one of the most important genes related to asthma phenotype and pathogenesis.
One of these cytokines plays an important role in the inflammatory response is IL13. IL13 is a factor stimulating Cell B to produce IgE; this has profound effects on epithelial cells, fibroblasts, smooth muscle cells, promoting airway restructure, involving in the production of mucus [51. Therefore, focusing on studying genes related to IL13, the authors found that the location of the IL13-coding genes related to the susceptibility of asthma on the 5p31 chromosome [52].
Functional polymorphism of IL13 analyzed, including polymorphism at the gene promoter located at 1112C / T altered transcription factors and help the appearance of amino acid polymorphism. Transforming a pair of bases in this position could lead to the substitution of arginine with glycine at this position 131. This will alter the affinity of IL13 at receptor IL13RA2 (IL13 binding in IL13RA2 receptor), increasing the functional activities through IL13RA1 and the molecular stability in plasma [18].
Polymorphisms in other genes encode the proteins of regulating Cells Th2 such as GATA3 Th2 cells (coding GATA- binding protein 3), TBX21 (coding Tbet, transcription factors for the development of Th1 cell growth), IL4 (encoding cytokines), ILR4 (receptor encoding IL4) and STAT6 (encoding the associated signal switch). Those are also associated with increasing susceptibility of asthma and asthma phenotype [53]. The evidence indicates that there may be a synergistic impact related to heredity rather than simple variations.
The genes involved in the pathogenesis of asthma can be divided into 5 groups [54]
The genes with direct expression under the influence of environmental factors include genes encoding components of the innate immune system [55] when the body is exposed to microorganisms that cause allergy immune responses.
For example, the CD14TLR4 gene encoding the lipopolysaccharide pathways (LPS) is important in innate immunity in asthma patients. Other environmental response genes include genes encoding removing enzymes (eg, glutathione S-transferase (GST) related to the exposure to redox, cigarette smoke and environment pollution).
Genes maintaining the integrity of the mucosal epithelial barrier send signaling from the epithelium to the immune system after the body exposure to environmental factors. Like filaggin in epithelial barrier, the appearance of genes encoding chitinase (eg CHIA, CHI3L1) plays an important role in allergic inflammation adjustment and increasing the expression and activity of epithelial cells, macrophages in patients with bronchial asthma. PCDH1 encoding protocadherin 1 is a member of the
family of cell adhesion molecules with the expression related to the bronchial epithelium and the increase of airway (bronchitis) reactivity.
Genes related to IL33 are identified by inflammatory genes and the genes with correlations in the entire chromosome, IL33 is produced by respiratory epithelial cells in order to response to the damage and stimulate Th2 cells to produce cytokines: IL4, IL5, IL13.
The immune response can be regulated by the genes, including conditioning through Th1/Th2 pathways and other function effects, eg IL6R activating the levels of lung inflammation [40]. Genes related to the tissue response in chronic inflammation like restructuring airways including: ADAM33 relates to the expression of fibrous and smooth muscle cells, PDE4D relates to smooth muscle cells and inflammatory cells, SMAD3 are proteins signaling in cells, activated by cytokine TGFβ -β transport growth factor.
Some genes do not directly relate to the asthma expression, but to the phenotypes, disease process such as acute asthma attacks rate, severity and coordinated obstruction. For example, genes which can be changed under the impact of vitamin D [56] or pollution particles with diameters of less than 10 mcg [57] related to the frequency of acute asthma attacks.
Asthma has diverse phenotypes, which may be due to the combination of the gene-related pathogenesis [48]. Studying on genes involved in the pathogenesis of asthma helps to classify the asthma phenotypes related to the pathogenesis and prognosis of treatment asthma on target.
The interactions between the gene - gene and gene- environment factors in Asthma
The interactions between genes
Like other complex genetic diseases, genes involved in asthma pathogenesis have the mutual interaction. The interactions between genes through the pathways of cytokine IL13/IL14 are typical. IL13 and IL14 in conjunction with receptor of IL4-α [IL4RA] causes the initial response to the division of Th2 cells. Then, IL13/IL4 produced by Th2 cells stimulates Cell B switches to produce IgE.
A case-control study in the Netherlands found the significant correlation between gene SNP S478P in IL4RA and 1112C/T in the polymorphism IL13. Individuals with genotypes containing the polymorphism in these two gene areas have asthma risk with 5 times higher than that of the general population (p=0.004). The data also shows that the interaction between IL4RA and IL13 increased the susceptibility of the individual to asthma [58]. A longitudinal study on 1120 children aged 9 to 11 indicates that a combination of genetic variations through IL14/IL13 pathways significantly relates to the development of allergies and asthma during childhood [59 ].
The intervention between genes and environment factors
The fact that individuals with the same genotype have different expression of phenotypes when interacting with various environmental factors may be due to out-of-gene expression. Out-of-gene expression - epigenomes are the gene expression changes which can be inherited without any change in the DNA sequence of the genome. The small chemical change will turn off or on the expression of the genes on the DNA.
Chemical tails are attached to DNA molecules by the methylation or wrapped so tightly around histone cores, so genes are not expressed. Under the impact of environmental factors: chemicals, diet,
medications, stress, or different development environments for each individual during pregnancy may stimulate DNA molecules open, enabling the genes expressed to synthesize protein molecules (phenotype modulation).
The researchers found that the out-of-gene expression associated with the asthma pathophysiology,
onset, clinical expression:
- Firstly, the rate of identical twins suffering from asthma was only about 50%, there should be other
elements causing asthma beside genes.
- Secondly, the complex interactions between genes and the environment related to asthma may reflect the changes of factors beyond genes. The study has found that the connection between pregnant women smoking during pregnancy and changes in the marker such as interleukin -1 receptor (IL-1RN) resistance factor, this increases children’s risk of asthma [60]. Smoking can change gene expression either directly or indirectly as a result of oxidative stress. Children of mothers who smoke during pregnancy have the reduction of methylation of AluYb8 in DNA sequences. The study also pointed out the SNP and in the sensitive locus in the chromosome region 17q21: ORMDL3 and GSDMB related to the onset of asthma in people exposed to tobacco [61].
- Thirdly, the gender differences in asthma is clearly noticed with higher incidence in boys before puberty, but higher in girls after puberty. At the same time, the children with asthma mothers have a higher risk of developing asthma than those with asthma fathers. This shows the possible impact of gender-related genes [22, 62-70].
The 17q21 variants strengthened the early asthma expression in children with respiratory infections before age of 2 years old.
CONCLUSION
Asthma is a syndrome with complex genetic characteristics. The study results demonstrated that asthma heredity is multi-genetic. One group of genes relates to the asthma susceptibility, allergies, severity and airway hypersusceptibility. Other groups of genes get involved in the pathways of medication response. Genetic studies of asthma showed a very important role of genes in asthma because it helps predict and classify asthma phenotypes and predict the medication response to personalize asthma treatment [71,72].
CONFLIT OF INTEREST
The authors declare non conflit of interest.
REFERENCE
1. Global initiative for Asthma (2016). <http://ginAsthma.org/wp-content/uploads/2016/04/GINA-2016-mainreport_tracked.pdf>.
2. S. T. Weiss, B. A. Raby và A. Rogers (2009). Asthma genetics and genomics 2009. Curr Opin Genet Dev, 19 (3), 279-282.
3. D. L. Duffy, N. G. Martin, D. Battistutta et al.(1990). Genetics of Asthma and hay fever in Australian twins. Am Rev Respir Dis, 142(6 Pt 1), 1351-1358.
4. J. R. Harris, P. Magnus, S. O. Samuelsen et al. (1997). No evidence for effects of family environment on Asthma. A retrospective study of Norwegian twins. Am J Respir Crit Care Med, 156(1), 43-49.
5. G. H. Koppelman, H. Los và D. S. Postma (1999). Genetic and environment in Asthma: the answer of twin studies. Eur Respir J, 13(1), 2-4.
6. M. M. Nieminen, J. Kaprio và M. Koskenvuo (1991). A population-based study of bronchial Asthma in adult twin pairs. Chest, 100(1), 70-75.
7. K. G. Tantisira, A. Damask, S. J. Szefler et al.(2012). Genome-wide association identifies the T gene as a novel Asthma pharmacogenetic locus. Am J Respir Crit Care Med, 185(12), 1286-1291.
8. M. E. March, P. M. Sleiman và H. Hakonarson (2011). The genetics of Asthma and allergic disorders. Discov Med, 11(56), 35-45.
9. Dominick Shaw, Michael Potelli và Ian Sayers (2014). Asthma. Handbook of Pharmacogenomics and Stratified Medicine, Elsevier Inc, 617-651.
10.S. T. Weiss, A. A. Litonjua, C. Lange et al.(2006). Overview of the pharmacogenetics of Asthma treatment. Pharmacogenomics J, 6(5),311-326.
11. M. E. Wechsler (2006). Managing Asthma in the 21st century: role of pharmacogenetics. Pediatr Ann, 35(9),660-662, 664-669.
12. K. G. Tantisira, S. Lake, E. S. Silverman et al.(2004). Corticosteroid pharmacogenetics: association of sequence variants in CRHR1 with improved lung function in Asthmatics treated with inhaled corticosteroids. Hum Mol Genet, 13(13), 1353-1359.
13. K. Tantisira and S. Weiss (2009). The pharmacogenetics of Asthma treatment. Current Allergy and Asthma Reports, 9(1), 10-17.
14. K. G. Tantisira, E. S. Silverman, T. J. Mariani et al. (2007). FCER2: a pharmacogenetic basis
for severe exacerbations in children with Asthma. J Allergy Clin Immunol, 120(6), 1285-1291.
15. Cook RA et Van der Veer A (1916). Human sensitisation. J Iminunol, 201-305.
16. J. W. Gerrard, P. Vickers và C. D. Gerrard (1976). The familial incidence of allergic disease. Ann Allergy, 36(1), 10-15.
17. L. R. Skadhauge, K. Christensen, K. O. Kyvik et al. (1999). Genetic and environmental influence on
Asthma: a population-based study of 11,688 Danish twin pairs. Eur Respir J, 13(1), 8-14.
18. John W. Holloway (2014). Genetics and Epigenetics of Allergic Diseases and Asthma. Middleton’s Allergy Principles and Practice, Elsevier Saunders, 1, 343-363.
19. P. Van Eerdewegh, R. D. Little, J. Dupuis et al. (2002). Association of the ADAM33 gene with Asthma and bronchial hyperresponsiveness. Nature, 418(6896), 426-430.
20. M. Allen, A. Heinzmann, E. Noguchi et al. (2003). Positional cloning of a novel gene influencing Asthma from chromosome 2q14. Nat Genet, 35(3), 258-263.
21. T. Laitinen, A. Polvi, P. Rydman et al. (2004). Characterization of a Common Susceptibility Locus for Asthma-Related Traits. Science, 304(5668), 300-304.
22. D. Nicolae, N. J. Cox, L. A. Lester et al. (2005). Fine mapping and positional candidate studies identify HLA-G as an Asthma susceptibility gene on chromosome 6p21. Am J Hum Genet, 76(2), 349-357.
23. Y. Zhang, N. I. Leaves, G. G. Anderson et al.(2003). Positional cloning of a quantitative trait locus on chromosome 13q14 that influences immunoglobulin E levels and Asthma. Nat Genet, 34(2), 181-186.
24. G. H. Koppelman, D. A. Meyers, T. D. Howard et al. (2009). Identification of PCDH1 as a novel susceptibility gene for bronchial hyperresponsiveness. Am J Respir Crit Care Med, 180(10), 929- 935.
25. C. N. A. Palmer, A. D. Irvine, A. Terron-Kwiatkowski et al. (2006). Common loss-of-function variants of the epidermal barrier protein filaggrin are a major predisposing factor for atopic dermatitis. Nat Genet, 38(4), 441-446.
26. E. Halapi, D. F. Gudbjartsson, G. M. Jonsdottir et al.(2010). A sequence variant on 17q21 is associated with age at onset and severity of Asthma. European Journal of Human Genetics, 18(8), 902-908.
27. T. F. Leung, H. Y. Sy, M. C. Ng et al. (2009). Asthma and atopy are associated with chromosome 17q21 markers in Chinese children. Allergy, 64(4), 621-628.
28. H. Wu, I. Romieu, J. J. Sienra-Monge et al. (2009). Genetic variation in ORM1-like 3 (ORMDL3) and gasdermin-like (GSDML) and childhood Asthma. Allergy, 64(4), 629-635.
29. C. L. M. Joseph, L. K. Williams, D. R. Ownby et al.(2006). Applying epidemiologic concepts of primary, secondary, and tertiary prevention to the elimination of racial disparities in Asthma. The Journal of allergy and clinical immunology, 117(2), 233-242.
30. A. J. Sandford, T. Chagani, S. Zhu et al. (2000). Polymorphisms in the IL4, IL4RA, and FCERIB genes and Asthma severity. J Allergy Clin Immunol, 106(1 Pt 1),135-140.
31. B. Beghe, S. Barton, S. Rorke et al. (2003). Polymorphisms in the interleukin-4 and interleukin-4 receptor alpha chain genes confer susceptibility to Asthma and atopy in a Caucasian population. Clin Exp Allergy, 33 (8), 1111-1117.
32. M. Kabesch, I. Tzotcheva, D. Carr et al. (2003). A complete screening of the IL4 gene: novel polymorphisms and their association with Asthma and IgE in childhood. J Allergy Clin Immunol, 112(5), 893-898.
33. P. S. Gao, X. Q. Mao, M. Baldini et al. (1999). Serum total IgE levels and CD14 on chromosome 5q31. Clin Genet, 56(2), 164-165.
34. T. F. Leung, N. L. Tang, Y. M. Sung et al. (2003). The C-159T polymorphism in the CD14 promoter is associated with serum total IgE concentration in atopic Chinese children. Pediatr Allergy Immunol, 14(4), 255-260.
35. M. Sharma, J. Batra, U. Mabalirajan et al. (2004). Suggestive evidence of association of C-159T functional polymorphism of the CD14 gene with atopic Asthma in northern and northwestern Indian populations. Immunogenetics, 56(7), 544-547.
36. A. C. Martin, I. A. Laing, S. K. Khoo et al. (2006). Acute Asthma in children: Relationships among CD14 and CC16 genotypes, plasma levels, and severity. Am J Respir Crit Care Med, 173(6), 617- 622.
37. M. F. Moffatt, M. Kabesch, L. Liang et al. (2007). Genetic variants regulating ORMDL3 expression contribute to the risk of childhood Asthma. Nature, 448(7152), 470-473.
38. D. G. Torgerson, E. J. Ampleford, G. Y. Chiu et al. (2011). Meta-analysis of genome-wide association studies of Asthma in ethnically diverse North American populations. Nat Genet, 43(9), 887-892.
39. G. M. Gauvreau , P. M. O'Byrne , L.-P. Boulet et al.(2014). Effects of an Anti-TSLP Antibody on AllergenInduced Asthmatic Responses. New England Journal of Medicine, 370(22), 2102-2110.
40. M. A. Ferreira, M. C. Matheson, D. L. Duffy et al.(2011). Identification of IL6R and chromosome
11q13.5 as risk loci for Asthma. Lancet, 378(9795),1006-1014.
41. T. Hirota, A. Takahashi, M. Kubo et al. (2011). Genomewide association study identifies three new susceptibility loci for adult Asthma in the Japanese population. Nat Genet, 43(9), 893-896.
42. D. F. Gudbjartsson, U. S. Bjornsdottir, E. Halapi et al.(2009). Sequence variants affecting eosinophil numbers associate with Asthma and myocardial infarction. Nat Genet, 41(3), 342-347.
43. X. Li, T. D. Howard, S. L. Zheng et al. (2010). Genomewide association study of Asthma identifies RAD50- IL13 and HLA-DR/DQ regions. J Allergy Clin Immunol, 125(2), 328-335 e311.
44. C. M. Dolan, K. E. Fraher, E. R. Bleecker et al.(2004). Design and baseline characteristics of the epidemiology and natural history of Asthma: Outcomes and Treatment Regimens (TENOR) study: a large cohort of patients with severe or difficult-totreat Asthma. Ann Allergy Asthma Immunol, 92(1),32-39.
45. T. Haselkorn, J. E. Fish, R. S. Zeiger et al. (2009). Consistently very poorly controlled Asthma, as defined by the impairment domain of the Expert Panel Report 3 guidelines, increases risk for future severe Asthma exacerbations in The Epidemiology and Natural History of Asthma: Outcomes and Treatment Regimens (TENOR) study. J Allergy Clin Immunol, 124(5), 895-902 e891-894.
46. X. Li, T. D. Howard, W. C. Moore et al. (2011). Importance of hedgehog interacting protein and other lung function genes in Asthma. J Allergy Clin Immunol, 127(6), 1457-1465.
47. S. E. Wenzel, S. Balzar, E. Ampleford et al.(2007). IL4R alpha mutations are associated with Asthma exacerbations and mast cell/IgE expression. Am J Respir Crit Care Med, 175(6),570-576.
48. W. C. Moore, D. A. Meyers, S. E. Wenzel et al.(2010). Identification of Asthma phenotypes using cluster analysis in the Severe Asthma Research Program. Am J Respir Crit Care Med, 181(4), 315- 323.
49. L. H. Slager RE, Moore WC, et al, (2011). Predictive model of severe atopic Asthma phenotypes using interleukin 4/13 pathway polymorphisms. Am J Respir Crit Care Med,183.
50. J. H. White, M. Chiano, M. Wigglesworth et al.(2008). Identification of a novel Asthma susceptibility gene on chromosome 1qter and its functional evaluation. Hum Mol Genet, 17(13), 1890-1903.
51. J. W. Holloway và G. H. Koppelman (2007). Identifying novel genes contributing to Asthma pathogenesis. Curr Opin Allergy Clin Immunol, 7(1), 69-74.
52. S. Hoffjan, D. Nicolae và C. Ober (2003). Association studies for Asthma and atopic diseases: a comprehensive review of the literature. Respiratory Research, 4(1), 14-14.
53. D. Vercelli (2008). Discovering susceptibility genes for Asthma and allergy. Nat Rev Immunol, 8(3), 169-182.
54. J. W. Holloway, I. A. Yang và S. T. Holgate (2010). Genetics of allergic disease. J Allergy Clin Immunol, 125(2 Suppl 2), S81-94.
55. I. A. Yang, S. Savarimuthu, S. T. Kim et al. (2007). Gene–environmental interaction in Asthma. Current Opinion in Allergy and Clinical Immunology, 7(1), 75-82.
56. R. Du, A. A. Litonjua, K. G. Tantisira et al.(2012). Genome-wide association study reveals class I MHC-restricted T cell-associated molecule gene (CRTAM) variants interact with vitamin D levels to affect Asthma exacerbations. J Allergy Clin Immunol, 129(2), 368-373, 373 e361-365.
57. C. Canova, C. Dunster, F. J. Kelly et al. (2012). PM10- induced hospital admissions for Asthma and chronic obstructive pulmonary disease: the modifying effect of individual characteristics. Epidemiology, 23(4), 607-615.
58. T. D. Howard, G. H. Koppelman, J. Xu et al. (2002). Gene-gene interaction in Asthma: IL4RA and IL13 in a Dutch population with Asthma. Am J Hum Genet, 70(1), 230-236.
59. M. Kabesch, M. Schedel, D. Carr et al. (2006). IL-4/IL-13 pathway genetics strongly influence serum IgE levels and childhood Asthma. J Allergy Clin Immunol, 117(2), 269-274.
60. R. A. Ramadas, A. Sadeghnejad, W. Karmaus et al.(2007). Interleukin-1R antagonist gene and pre-natal smoke exposure are associated with childhood Asthma. The European respiratory journal: official journal of the European Society for Clinical Respiratory Physiology, 29(3), 502-508.
61. E. Bouzigon , E. Corda , H. Aschard et al. (2008). Effect of 17q21 Variants and Smoking Exposure in
Early-Onset Asthma. New England Journal of Medicine, 359(19), 1985-1994.
62. A. A. Litonjua, V. J. Carey, H. A. Burge et al. (1998). Parental history and the risk for childhood Asthma. Does mother confer more risk than father? Am J Respir Crit Care Med, 158(1), 176-181.
63. W. O. Cookson, R. P. Young, A. J. Sandford et al.(1992). Maternal inheritance of atopic IgE responsiveness on chromosome 11q. Lancet, 340(8816), 381- 384.
64. M. R. Sears, M. D. Holdaway, E. M. Flannery et al.(1996). Parental and neonatal risk factors for atopy, airway hyper-responsiveness, and Asthma. Archives of Disease in Childhood, 75(5), 392-398.
65. H. Lin, T. R. Mosmann, L. Guilbert et al. (1993). Synthesis of T helper 2-type cytokines at the maternal-fetal interface. J Immunol, 151(9), 4562-4573.
66. J. M. Drazen, E. K. Silverman và T. H. Lee (2000). Heterogeneity of therapeutic responses in asthma. Br Med Bull, 56 (4), 1054-1070.
67. E. Reihsaus, M. Innis, N. MacIntyre và cộng sự(1993). Mutations in the gene encoding for the beta 2-adrenergic receptor in normal and asthmatic subjects. Am J Respir Cell Mol Biol, 8 (3), 334-339.
68. F. D. Martinez, P. E. Graves, M. Baldini và cộng sự (1997). Association between genetic polymorphisms of the beta2-adrenoceptor and response to albuterol in children with and without a history of wheezing. Journal of Clinical Investigation, 100 (12), 3184-3188.
69. D. R. Taylor, R. J. Hancox, W. McRae và cộng sự (2000). The influence of polymorphism at position 16 of the beta2-adrenoceptor on the development of tolerance to beta-agonist. J Asthma, 37 (8), 691-700.
70. E. Israel, J. M. Drazen, S. B. Liggett và cộng sự (2000). The effect of polymorphisms of the beta(2)- adrenergic receptor on the response to regular use of albuterol in asthma. Am J Respir Crit Care Med, 162 (1), 75-80.
71. S. Duong-Quy. Asthma and chronic sinusitis: one disease for two organs. J Fran Viet Pneu 2016;20(7):1-2.
72. H. Duong Thi Ly, N. Pham Thi Hong, T. Vu Thi, H. Nguyen Thi Bich, H. Le Thi Minh, S. Duong Quy. The frequency of FCER2 genotype distributions related to corticosteroid response in asthma patients treated at the National Hospital of Paediatrics. J Fran Viet Pneu 2016;20(7):54- 59.
73. R.E. Slager, Recent developments in the genetics of asthma susceptibility and severity, European Respiratory monograph, Number 51, March 2011, Pages 82-96.
FIGURE
REFERENCE
1. Global initiative for Asthma (2016). <http://ginAsthma.org/wp-content/uploads/2016/04/GINA-2016-mainreport_tracked.pdf>.
2. S. T. Weiss, B. A. Raby và A. Rogers (2009). Asthma genetics and genomics 2009. Curr Opin Genet Dev, 19 (3), 279-282.
3. D. L. Duffy, N. G. Martin, D. Battistutta et al.(1990). Genetics of Asthma and hay fever in Australian twins. Am Rev Respir Dis, 142(6 Pt 1), 1351-1358.
4. J. R. Harris, P. Magnus, S. O. Samuelsen et al. (1997). No evidence for effects of family environment on Asthma. A retrospective study of Norwegian twins. Am J Respir Crit Care Med, 156(1), 43-49.
5. G. H. Koppelman, H. Los và D. S. Postma (1999). Genetic and environment in Asthma: the answer of twin studies. Eur Respir J, 13(1), 2-4.
6. M. M. Nieminen, J. Kaprio và M. Koskenvuo (1991). A population-based study of bronchial Asthma in adult twin pairs. Chest, 100(1), 70-75.
7. K. G. Tantisira, A. Damask, S. J. Szefler et al.(2012). Genome-wide association identifies the T gene as a novel Asthma pharmacogenetic locus. Am J Respir Crit Care Med, 185(12), 1286-1291.
8. M. E. March, P. M. Sleiman và H. Hakonarson (2011). The genetics of Asthma and allergic disorders. Discov Med, 11(56), 35-45.
9. Dominick Shaw, Michael Potelli và Ian Sayers (2014). Asthma. Handbook of Pharmacogenomics and Stratified Medicine, Elsevier Inc, 617-651.
10.S. T. Weiss, A. A. Litonjua, C. Lange et al.(2006). Overview of the pharmacogenetics of Asthma treatment. Pharmacogenomics J, 6(5),311-326.
11. M. E. Wechsler (2006). Managing Asthma in the 21st century: role of pharmacogenetics. Pediatr Ann, 35(9),660-662, 664-669.
12. K. G. Tantisira, S. Lake, E. S. Silverman et al.(2004). Corticosteroid pharmacogenetics: association of sequence variants in CRHR1 with improved lung function in Asthmatics treated with inhaled corticosteroids. Hum Mol Genet, 13(13), 1353-1359.
13. K. Tantisira and S. Weiss (2009). The pharmacogenetics of Asthma treatment. Current Allergy and Asthma Reports, 9(1), 10-17.
14. K. G. Tantisira, E. S. Silverman, T. J. Mariani et al. (2007). FCER2: a pharmacogenetic basis
for severe exacerbations in children with Asthma. J Allergy Clin Immunol, 120(6), 1285-1291.
15. Cook RA et Van der Veer A (1916). Human sensitisation. J Iminunol, 201-305.
16. J. W. Gerrard, P. Vickers và C. D. Gerrard (1976). The familial incidence of allergic disease. Ann Allergy, 36(1), 10-15.
17. L. R. Skadhauge, K. Christensen, K. O. Kyvik et al. (1999). Genetic and environmental influence on
Asthma: a population-based study of 11,688 Danish twin pairs. Eur Respir J, 13(1), 8-14.
18. John W. Holloway (2014). Genetics and Epigenetics of Allergic Diseases and Asthma. Middleton’s Allergy Principles and Practice, Elsevier Saunders, 1, 343-363.
19. P. Van Eerdewegh, R. D. Little, J. Dupuis et al. (2002). Association of the ADAM33 gene with Asthma and bronchial hyperresponsiveness. Nature, 418(6896), 426-430.
20. M. Allen, A. Heinzmann, E. Noguchi et al. (2003). Positional cloning of a novel gene influencing Asthma from chromosome 2q14. Nat Genet, 35(3), 258-263.
21. T. Laitinen, A. Polvi, P. Rydman et al. (2004). Characterization of a Common Susceptibility Locus for Asthma-Related Traits. Science, 304(5668), 300-304.
22. D. Nicolae, N. J. Cox, L. A. Lester et al. (2005). Fine mapping and positional candidate studies identify HLA-G as an Asthma susceptibility gene on chromosome 6p21. Am J Hum Genet, 76(2), 349-357.
23. Y. Zhang, N. I. Leaves, G. G. Anderson et al.(2003). Positional cloning of a quantitative trait locus on chromosome 13q14 that influences immunoglobulin E levels and Asthma. Nat Genet, 34(2), 181-186.
24. G. H. Koppelman, D. A. Meyers, T. D. Howard et al. (2009). Identification of PCDH1 as a novel susceptibility gene for bronchial hyperresponsiveness. Am J Respir Crit Care Med, 180(10), 929- 935.
25. C. N. A. Palmer, A. D. Irvine, A. Terron-Kwiatkowski et al. (2006). Common loss-of-function variants of the epidermal barrier protein filaggrin are a major predisposing factor for atopic dermatitis. Nat Genet, 38(4), 441-446.
26. E. Halapi, D. F. Gudbjartsson, G. M. Jonsdottir et al.(2010). A sequence variant on 17q21 is associated with age at onset and severity of Asthma. European Journal of Human Genetics, 18(8), 902-908.
27. T. F. Leung, H. Y. Sy, M. C. Ng et al. (2009). Asthma and atopy are associated with chromosome 17q21 markers in Chinese children. Allergy, 64(4), 621-628.
28. H. Wu, I. Romieu, J. J. Sienra-Monge et al. (2009). Genetic variation in ORM1-like 3 (ORMDL3) and gasdermin-like (GSDML) and childhood Asthma. Allergy, 64(4), 629-635.
29. C. L. M. Joseph, L. K. Williams, D. R. Ownby et al.(2006). Applying epidemiologic concepts of primary, secondary, and tertiary prevention to the elimination of racial disparities in Asthma. The Journal of allergy and clinical immunology, 117(2), 233-242.
30. A. J. Sandford, T. Chagani, S. Zhu et al. (2000). Polymorphisms in the IL4, IL4RA, and FCERIB genes and Asthma severity. J Allergy Clin Immunol, 106(1 Pt 1),135-140.
31. B. Beghe, S. Barton, S. Rorke et al. (2003). Polymorphisms in the interleukin-4 and interleukin-4 receptor alpha chain genes confer susceptibility to Asthma and atopy in a Caucasian population. Clin Exp Allergy, 33 (8), 1111-1117.
32. M. Kabesch, I. Tzotcheva, D. Carr et al. (2003). A complete screening of the IL4 gene: novel polymorphisms and their association with Asthma and IgE in childhood. J Allergy Clin Immunol, 112(5), 893-898.
33. P. S. Gao, X. Q. Mao, M. Baldini et al. (1999). Serum total IgE levels and CD14 on chromosome 5q31. Clin Genet, 56(2), 164-165.
34. T. F. Leung, N. L. Tang, Y. M. Sung et al. (2003). The C-159T polymorphism in the CD14 promoter is associated with serum total IgE concentration in atopic Chinese children. Pediatr Allergy Immunol, 14(4), 255-260.
35. M. Sharma, J. Batra, U. Mabalirajan et al. (2004). Suggestive evidence of association of C-159T functional polymorphism of the CD14 gene with atopic Asthma in northern and northwestern Indian populations. Immunogenetics, 56(7), 544-547.
36. A. C. Martin, I. A. Laing, S. K. Khoo et al. (2006). Acute Asthma in children: Relationships among CD14 and CC16 genotypes, plasma levels, and severity. Am J Respir Crit Care Med, 173(6), 617- 622.
37. M. F. Moffatt, M. Kabesch, L. Liang et al. (2007). Genetic variants regulating ORMDL3 expression contribute to the risk of childhood Asthma. Nature, 448(7152), 470-473.
38. D. G. Torgerson, E. J. Ampleford, G. Y. Chiu et al. (2011). Meta-analysis of genome-wide association studies of Asthma in ethnically diverse North American populations. Nat Genet, 43(9), 887-892.
39. G. M. Gauvreau , P. M. O'Byrne , L.-P. Boulet et al.(2014). Effects of an Anti-TSLP Antibody on AllergenInduced Asthmatic Responses. New England Journal of Medicine, 370(22), 2102-2110.
40. M. A. Ferreira, M. C. Matheson, D. L. Duffy et al.(2011). Identification of IL6R and chromosome
11q13.5 as risk loci for Asthma. Lancet, 378(9795),1006-1014.
41. T. Hirota, A. Takahashi, M. Kubo et al. (2011). Genomewide association study identifies three new susceptibility loci for adult Asthma in the Japanese population. Nat Genet, 43(9), 893-896.
42. D. F. Gudbjartsson, U. S. Bjornsdottir, E. Halapi et al.(2009). Sequence variants affecting eosinophil numbers associate with Asthma and myocardial infarction. Nat Genet, 41(3), 342-347.
43. X. Li, T. D. Howard, S. L. Zheng et al. (2010). Genomewide association study of Asthma identifies RAD50- IL13 and HLA-DR/DQ regions. J Allergy Clin Immunol, 125(2), 328-335 e311.
44. C. M. Dolan, K. E. Fraher, E. R. Bleecker et al.(2004). Design and baseline characteristics of the epidemiology and natural history of Asthma: Outcomes and Treatment Regimens (TENOR) study: a large cohort of patients with severe or difficult-totreat Asthma. Ann Allergy Asthma Immunol, 92(1),32-39.
45. T. Haselkorn, J. E. Fish, R. S. Zeiger et al. (2009). Consistently very poorly controlled Asthma, as defined by the impairment domain of the Expert Panel Report 3 guidelines, increases risk for future severe Asthma exacerbations in The Epidemiology and Natural History of Asthma: Outcomes and Treatment Regimens (TENOR) study. J Allergy Clin Immunol, 124(5), 895-902 e891-894.
46. X. Li, T. D. Howard, W. C. Moore et al. (2011). Importance of hedgehog interacting protein and other lung function genes in Asthma. J Allergy Clin Immunol, 127(6), 1457-1465.
47. S. E. Wenzel, S. Balzar, E. Ampleford et al.(2007). IL4R alpha mutations are associated with Asthma exacerbations and mast cell/IgE expression. Am J Respir Crit Care Med, 175(6),570-576.
48. W. C. Moore, D. A. Meyers, S. E. Wenzel et al.(2010). Identification of Asthma phenotypes using cluster analysis in the Severe Asthma Research Program. Am J Respir Crit Care Med, 181(4), 315- 323.
49. L. H. Slager RE, Moore WC, et al, (2011). Predictive model of severe atopic Asthma phenotypes using interleukin 4/13 pathway polymorphisms. Am J Respir Crit Care Med,183.
50. J. H. White, M. Chiano, M. Wigglesworth et al.(2008). Identification of a novel Asthma susceptibility gene on chromosome 1qter and its functional evaluation. Hum Mol Genet, 17(13), 1890-1903.
51. J. W. Holloway và G. H. Koppelman (2007). Identifying novel genes contributing to Asthma pathogenesis. Curr Opin Allergy Clin Immunol, 7(1), 69-74.
52. S. Hoffjan, D. Nicolae và C. Ober (2003). Association studies for Asthma and atopic diseases: a comprehensive review of the literature. Respiratory Research, 4(1), 14-14.
53. D. Vercelli (2008). Discovering susceptibility genes for Asthma and allergy. Nat Rev Immunol, 8(3), 169-182.
54. J. W. Holloway, I. A. Yang và S. T. Holgate (2010). Genetics of allergic disease. J Allergy Clin Immunol, 125(2 Suppl 2), S81-94.
55. I. A. Yang, S. Savarimuthu, S. T. Kim et al. (2007). Gene–environmental interaction in Asthma. Current Opinion in Allergy and Clinical Immunology, 7(1), 75-82.
56. R. Du, A. A. Litonjua, K. G. Tantisira et al.(2012). Genome-wide association study reveals class I MHC-restricted T cell-associated molecule gene (CRTAM) variants interact with vitamin D levels to affect Asthma exacerbations. J Allergy Clin Immunol, 129(2), 368-373, 373 e361-365.
57. C. Canova, C. Dunster, F. J. Kelly et al. (2012). PM10- induced hospital admissions for Asthma and chronic obstructive pulmonary disease: the modifying effect of individual characteristics. Epidemiology, 23(4), 607-615.
58. T. D. Howard, G. H. Koppelman, J. Xu et al. (2002). Gene-gene interaction in Asthma: IL4RA and IL13 in a Dutch population with Asthma. Am J Hum Genet, 70(1), 230-236.
59. M. Kabesch, M. Schedel, D. Carr et al. (2006). IL-4/IL-13 pathway genetics strongly influence serum IgE levels and childhood Asthma. J Allergy Clin Immunol, 117(2), 269-274.
60. R. A. Ramadas, A. Sadeghnejad, W. Karmaus et al.(2007). Interleukin-1R antagonist gene and pre-natal smoke exposure are associated with childhood Asthma. The European respiratory journal: official journal of the European Society for Clinical Respiratory Physiology, 29(3), 502-508.
61. E. Bouzigon , E. Corda , H. Aschard et al. (2008). Effect of 17q21 Variants and Smoking Exposure in Early-Onset Asthma. New England Journal of Medicine, 359(19), 1985-1994.
62. A. A. Litonjua, V. J. Carey, H. A. Burge et al. (1998). Parental history and the risk for childhood Asthma. Does mother confer more risk than father? Am J Respir Crit Care Med, 158(1), 176-181.
63. W. O. Cookson, R. P. Young, A. J. Sandford et al.(1992). Maternal inheritance of atopic IgE responsiveness on chromosome 11q. Lancet, 340(8816), 381- 384.
64. M. R. Sears, M. D. Holdaway, E. M. Flannery et al.(1996). Parental and neonatal risk factors for atopy, airway hyper-responsiveness, and Asthma. Archives of Disease in Childhood, 75(5), 392-398.
65. H. Lin, T. R. Mosmann, L. Guilbert et al. (1993). Synthesis of T helper 2-type cytokines at the maternal-fetal interface. J Immunol, 151(9), 4562-4573.
66. J. M. Drazen, E. K. Silverman và T. H. Lee (2000). Heterogeneity of therapeutic responses in asthma. Br Med Bull, 56 (4), 1054-1070.
67. E. Reihsaus, M. Innis, N. MacIntyre và cộng sự(1993). Mutations in the gene encoding for the beta 2-adrenergic receptor in normal and asthmatic subjects. Am J Respir Cell Mol Biol, 8 (3), 334-339.
68. F. D. Martinez, P. E. Graves, M. Baldini và cộng sự (1997). Association between genetic polymorphisms of the beta2-adrenoceptor and response to albuterol in children with and without a history of wheezing. Journal of Clinical Investigation, 100 (12), 3184-3188.
69. D. R. Taylor, R. J. Hancox, W. McRae và cộng sự (2000). The influence of polymorphism at position 16 of the beta2-adrenoceptor on the development of tolerance to beta-agonist. J Asthma, 37 (8), 691-700.
70. E. Israel, J. M. Drazen, S. B. Liggett và cộng sự (2000). The effect of polymorphisms of the beta(2)- adrenergic receptor on the response to regular use of albuterol in asthma. Am J Respir Crit Care Med, 162 (1), 75-80.
71. S. Duong-Quy. Asthma and chronic sinusitis: one disease for two organs. J Fran Viet Pneu 2016;20(7):1-2.
72. H. Duong Thi Ly, N. Pham Thi Hong, T. Vu Thi, H. Nguyen Thi Bich, H. Le Thi Minh, S. Duong Quy. The frequency of FCER2 genotype distributions related to corticosteroid response in asthma patients treated at the National Hospital of Paediatrics. J Fran Viet Pneu 2016;20(7):54- 59.
73. R.E. Slager, Recent developments in the genetics of asthma susceptibility and severity, European Respiratory monograph, Number 51, March 2011, Pages 82-96.